Stacking Up Against Sequencing
Last week we compared results from qPCR vs. 16S, demonstrating Branchpoint assays provided superior taxonomic resolution, better accuracy, and greater sensitivity.
But can qPCR-based microbiome profiling really compete with deep metagenomic sequencing? We went head-to-head against standard industry approaches so you can judge for yourself.
qPCR vs. Shotgun Sequencing
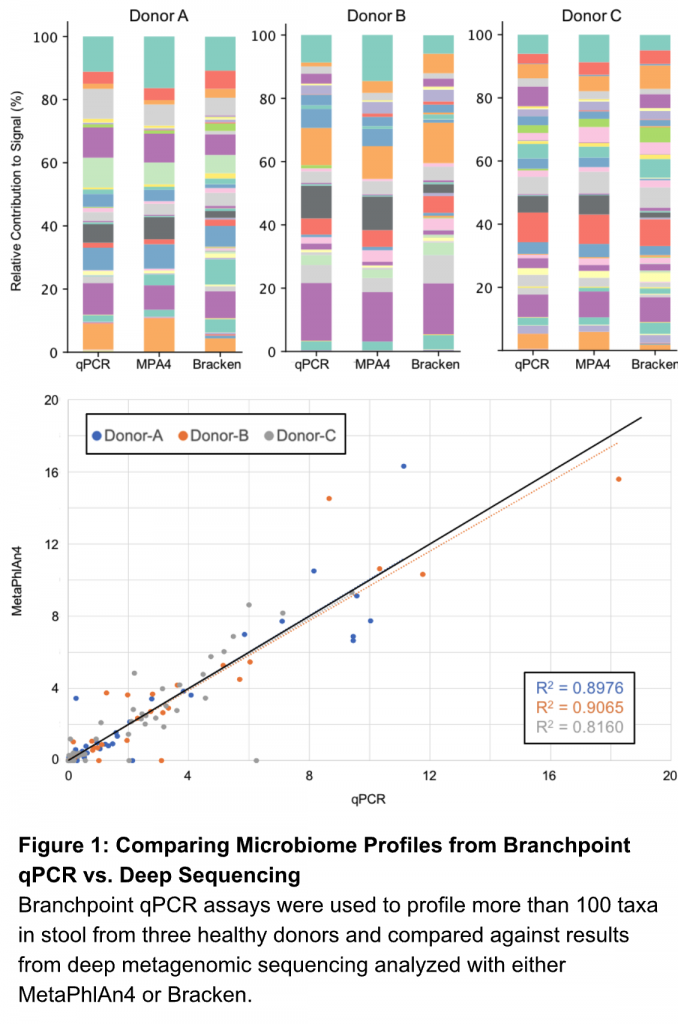
We compared more than 100 of our species-specific assays against deep shotgun metagenomic sequencing – 300M paired-end reads – on stool samples from 3 healthy human donors.
Let’s see how qPCR stacks up (Figure 1).
Our assays provided absolute abundance (genome equivalents) that mirrored results from the most widely cited bioinformatics analysis pipelines (MetaPhlAn or Kracken2/Bracken). The relative contributions from each species was in agreement with sequencing results analyzed with either bioinformatics pipeline. In fact, linear regression analysis for all 3 samples demonstrated that our qPCR-based community profiles corresponded better with MetaPhlAn4 and Bracken results than the sequencing methods compared against each other.
Modern design captures meaningful diversity
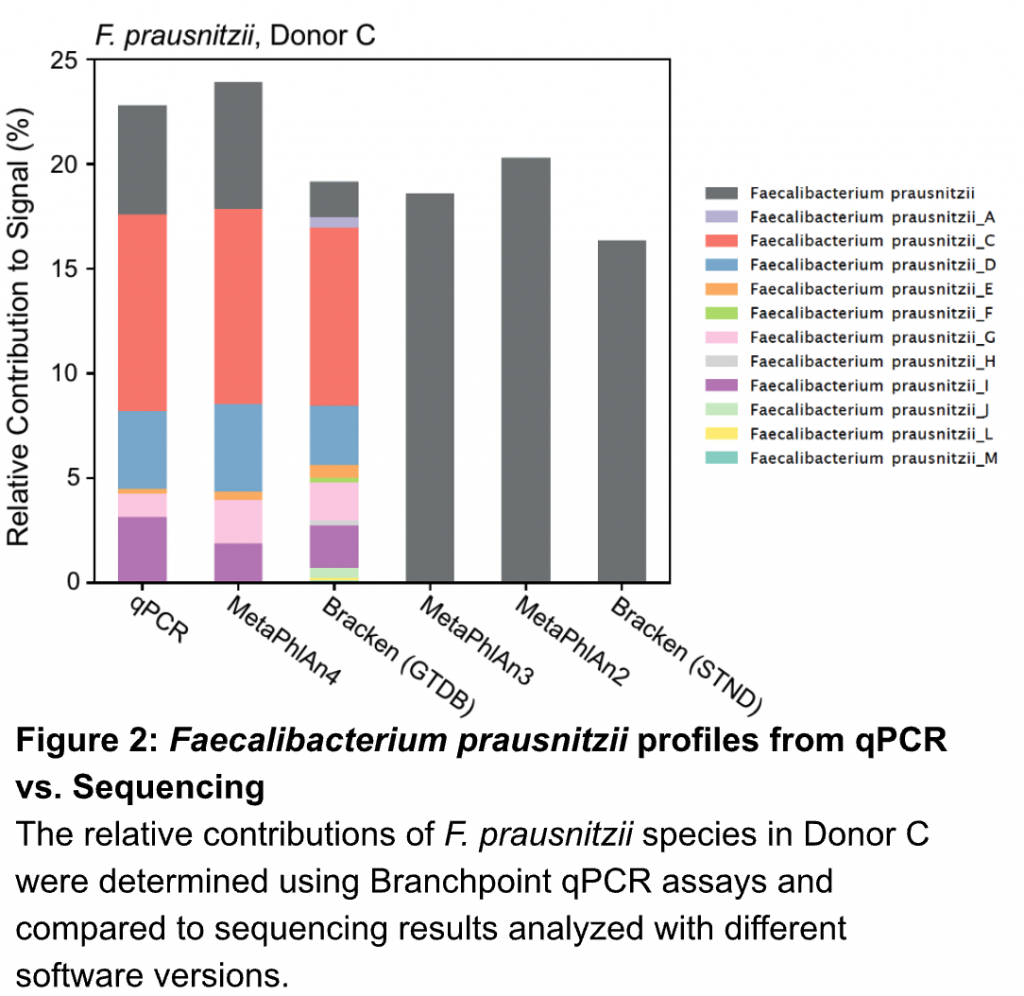
We design our assays to go beyond canonical (often outdated) species designations to provide superior taxonomic resolution. For example, the group of organisms historically known as Faecalibacterium prausnitzii actually contains many distinct species. Accordingly, we integrated these modern insights into our assays to capture this important biodiversity.
Our assays revealed many of these distinct F. prausnitzii within one of our healthy donors, recapitulating the results found with deep metagenomic sequencing using modern analysis tools (MetaPhlAn4)(Figure 2).
Older versions of bioinformatic pipelines (MetaPhlAn3) or alternative databases (Bracken – STND) couldn’t even provide this fine scale resolution, often obscuring relevant information by lumping biologically distinct organisms into one group. Our assays provide meaningful taxonomic resolution so you can see the signal from the noise.
Counting needles in the haystack to reduce false negatives
Metagenomic approaches characterize the most abundant sequences in a community leaving rare organisms below detection. Accordingly, the targeted nature of PCR offers superior sensitivity.
Across the three samples we observed 40 cases across 330 observations (12%) where our qPCR assays detected a species that sequencing missed. Ranging between 1 and 6,211 genome copies within a complex sample of 1.7 million genomes, these rare organisms were ignored as false negatives, despite 300M reads of sequencing depth.
Quality Counts: Sensitivity with Specificity
While providing comparable community profiles, qPCR also offers some significant advantages. Absolute quantification overcomes limitations inherent to compositional data sets from sequencing, and the superior sensitivity of qPCR counts alleviates the issue of zero inflation in statistical analysis. Assays designed with taxonomic specificity also streamline analysis to deliver precise profiles faster and easier than sequencing.
These attributes make qPCR-based microbiome profiling a better option for many applications. Next week we’ll share a few examples of how qPCR-based profiling can open up new opportunities for your microbiome analysis.
