How Reliable is your Microbiome Data?
It’s hard to advance any field without accurate data, and that’s especially true when it comes to the human microbiome.
Understanding the growing need for standardization, the US National Institute of Standards and Technology (NIST) – the maker and keeper of commercial standards and references (Did you know there are NIST standards for cigarettes, peanut butter, and slurried spinach?) – is in the process of creating human stool microbiome reference material and asking the question: how reliably can we measure the microbiome?
In doing so, they have published the scariest two figures for anyone trying to capture the biological reality of microbiome samples.
The Set-up
NIST produced reference material to be sequenced by laboratories around the World using their own in-house sample preparation methods. The material consisted of homogenized stool donations from multiple individuals and two mock communities of known DNA. Labs were asked to process these samples using their standard operating procedures and produce raw sequencing data using either 16S amplicon sequencing or whole shotgun metagenomic sequencing. The raw data was returned to NIST, who then analyzed the results in a standardized manner.
The Results
Given the compositional nature of sequencing data (more on this topic to come!), NIST assessed the ratios between groups of organisms… and the results are wild (bioRxiv preprint here).
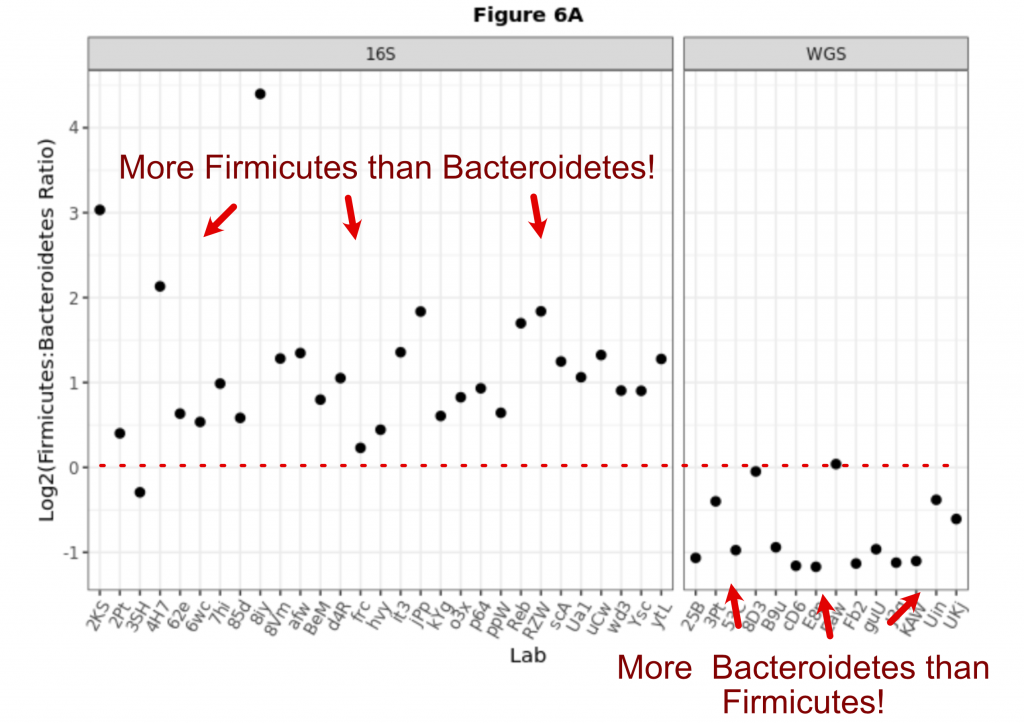
Borrowed from the preprint, Figure 6A, the first figure shows that different methods give you fundamentally different answers! If the lab used 16S amplicons then Firmicutes were more abundant than Bacteroidetes. But if a lab used shotgun sequencing it was the opposite! Early results using 16S suggested the ratio of Firmicutes to Bacteroidetes was indicative of a healthy or obese microbiome and fueled a decade of follow-up studies. It turns out, this ratio might have been a mere artifact of the sequencing methodology. But as this shows, even for the same sample, labs cannot not agree!
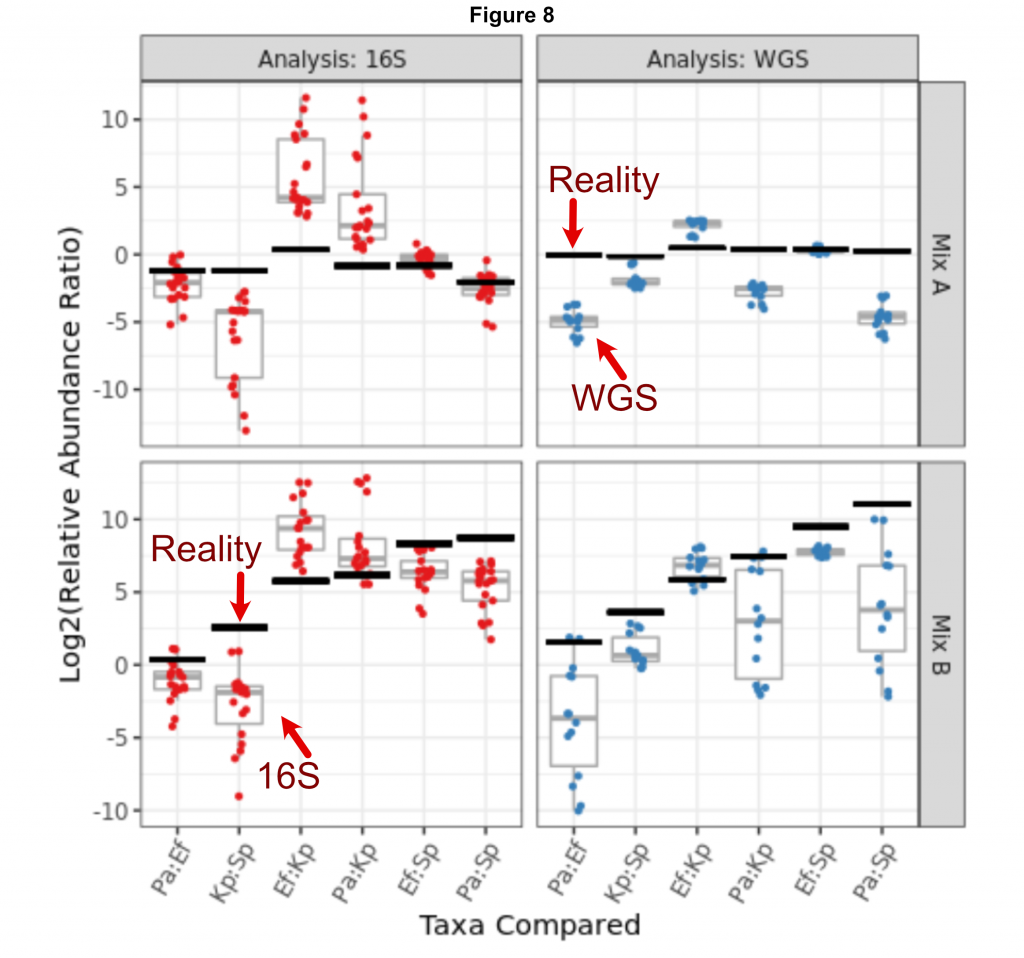
Just as troubling, labs struggled to produce the same answer even when using simple defined mock communities, again borrowed from the preprint Figure 8, here using two defined mock communities – one with four strains at equal abundance and the other staggered – split between 16S in red and shotgun sequencing in blue. The black line denotes the expected ratio between two species within the mock communities and each dot is a result from a different lab. Only in a few instances does the consensus between labs match the expected values. So when a lab asks either sequencing method to determine which microbes are present, often the result does not reflect biological reality.
How much confidence do you have in the reliability of your sequencing?
Branchpoint Biosciences Quantitative Microbiome Profiling (qMP™)
The microbiome field needs a technology it can rely on to capture biological reality. Having a standardized method for processing material and producing sequencing inputs is only part of the battle. By harnessing quantifiably, reproducible microbiome profiles it is possible to definitively know which microbes are vital to your signal and translate that information into action.
